Project members
Project
Approximately one in ten men suffers from infertility most often resulting from an abnormal spermatogenesis. The cause, whether genetic or environmental, remains often unknown, with ~50% of male infertility defined as ‘idiopathic’. Spermatogenesis, the process by which spermatozoa develop from immature germ cells in the seminiferous tubules of the testis, is associated with a highly dynamic genetic program and extensive chromatin changes.
Our group studies spermatogenesis at the gene and the chromatin levels to identify and characterize novel regulators/pathways required for sperm differentiation and male fertility. We are particularly interested in genes carried by the sex chromosomes and expressed in male germ cells since their deregulation impairs male fertility. Our goal is to produce fundamental knowledge on the molecular mechanisms controlling spermatogenesis in order to better understand the genetic and epigenetic causes of male infertility.
Our project also aims at studying male gamete epigenome in health and disease. Indeed it is now well-known that, upon fertilization, the sperm cell contributes to the embryo with more than its DNA: epigenetic information (chromatin "code", non-coding RNAs, DNA modifications) is also transmitted to the embryo and could affect embryo development and offspring health, in case of deregulation. A better characterization of the molecular mechanisms controlling the establishment of paternal gamete epigenome is therefore crucial.
To address these questions, we combine in vivo and in vitro models and use molecular and cell biology, biochemistry and reproductive biology techniques. In particular we perform large-scale (‘omics) analyses such as transcriptomics, epigenomics and proteomics.
Selected references
Moretti et al. Cell Death & Diff 2017
Cocquet et al. PLoS Genet 2012
News
¤ January 2025 Publication of our article in Communications Biology
¤ November 2024: Congratulations @ Julie Cocquet on her success at the Inserm Research Director competitive recruitment.
¤ November 2024: Welcome to Gabrielle Maillard, our new Master 2 student !
¤ February 2024: Congratulations @ Alberto de la Iglesia and Carina Prip-Buus for the PIC (Inter Cochin Project) funding they obtained.
¤ October 2023: Congratulations @ Alberto de la Iglesia on securing a 3-year postdoctoral fellowship from the FRM.
¤ October 2023: Congratulations @ Manon Coulée on defending and obtaining her PhD
¤ June 2023: Congratulations @ Manon Coulée on securing a 6-month postdoctoral transition fellowship from the Labex Who Am I ?
¤ April 2023 Publication of our article in EMBO Reports

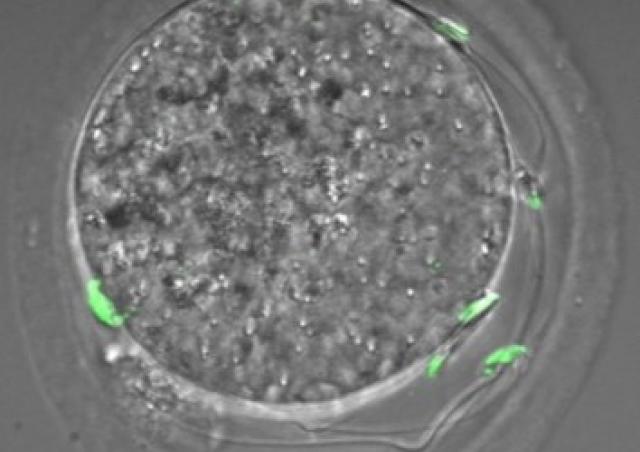
From Moretti et al. 2017. Localization of the histone methyl transferase DOT1L (green) in mouse male germ cells at different stages of their development.
DAPI (in blue) stains nuclei and Lectin-PNA (in red) marks the acrosomes.

Figure: From Cocquet et al. PLoS Genet 2012. Localization of SLX and SLY proteins in spermatids. SLX and SLY proteins (in green, top and bottom panel respectively) co-localize with the X or Y chromosome (in yellow, X or Y paint) and with Speer gene cluster (in red, DNA FISH). DAPI (blue) was used to stain nuclei.