Margottin-Goguet group composition
Projects
The molecular battle at stake between the host cell and the virus is at the heart of the ongoing research of our team. Our general objectives are to better understand the role of viral accessory proteins and to identify and characterize host restrictions factors. Understanding how the virus is blocked at different steps could lead to the proposal of new antiviral therapies.
Our main lines of research are:
-The search for host proteins targeted by viral auxiliary proteins from HIV-1/HIV-2/SIV. Recently, we have discovered that HUSH, a complex that is involved in the repression of gene expression, is degraded in the presence of the HIV-2/SIVsmm Vpx protein (Chougui et al, Nature microbio 2018)
-The study of the mechanism of viral antagonism by divergent lentiviral proteins. Vpx induces the degradation of two host factors: SAMHD1 and HUSH. In both cases, degradation relies on the hijacking of the DCAF1 adaptor of a Cullin 4A-based ubiquitin ligase. Nonetheless, our recent results show that differences exist between SAMHD1 and HUSH antagonism by Vpx (Martin*, Matkovic* et al, plos path 2021)
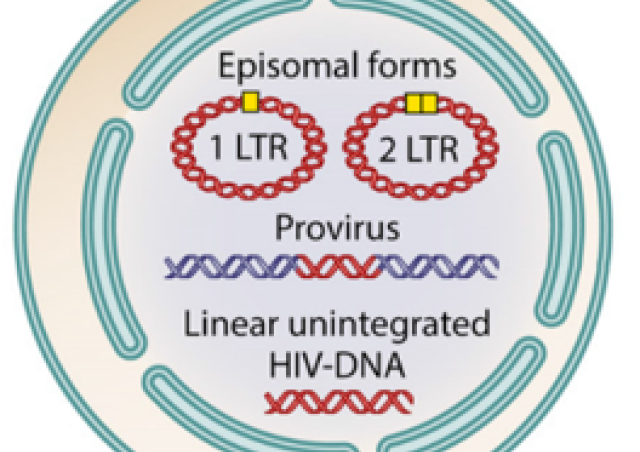
-The study of the mechanism of action of host restriction factors. Our team has contributed to characterize the mechanisms of action of SAMHD1 and HUSH. SAMHD1 is a dNTP hydrolase, which depletes nucleotides; in turn SAMHD1 inhibits the synthesis of viral DNA in non-dividing cells (Lahouassa et al, Nature Immunology 2012). HUSH represses expression of the viral genome integrated into the host genome, both at the epigenetic and post-transcriptional levels (Chougui et al, Nature microbio 2018; Matkovic et al, Nature com 2022).
Selected contributions of the team to the HIV field
- Découverte du mécanisme de dégradation du CD4 par Vpu. Premier exemple de détournement d’une ubiquitine ligase par une protéine du VIH (deuxième exemple dans le monde des pathogènes).
A novel human WD protein, h-beta TrCp, that interacts with HIV-1 Vpu connects CD4 to the ER degradation pathway through an F-box motif.
Margottin F, Bour SP, Durand H, Selig L, Benichou S, Richard V, Thomas D, Strebel K, Benarous R. Mol Cell. 1998 Mar;1(4):565-74. doi: 10.1016/s1097-2765(00)80056-8.
- Découverte du mécanisme d’action de la protéine virale Vpr de VIH-1 et première démonstration que Vpr de HIV-1 et Vpx de VIH-2 utilisent la même ubiquitine ligase (le point de départ de la découverte de certains facteurs de restriction).
HIV1 Vpr arrests the cell cycle by recruiting DCAF1/VprBP, a receptor of the Cul4-DDB1 ubiquitin ligase.
Le Rouzic E, Belaïdouni N, Estrabaud E, Morel M, Rain JC, Transy C, Margottin-Goguet F. Cell Cycle. 2007 Jan 15;6(2):182-8. doi: 10.4161/cc.6.2.3732.
- Découverte du mécanisme d’action de SAMHD1 dans les cellules myéloïdes.
SAMHD1 restricts the replication of human immunodeficiency virus type 1 by depleting the intracellular pool of deoxynucleoside triphosphates.
Lahouassa H, Daddacha W, Hofmann H, Ayinde D, Logue EC, Dragin L, Bloch N, Maudet C, Bertrand M, Gramberg T, Pancino G, Priet S, Canard B, Laguette N, Benkirane M, Transy C, Landau NR, Kim B, Margottin-Goguet F. Nat Immunol. 2012 Feb 12;13(3):223-228. doi: 10.1038/ni.2236.
- Démonstration que la restriction SAMHD1 n’est pas impliquée dans le bloc interféron qui diminue l’infection par le VIH-1.
Interferon block to HIV-1 transduction in macrophages despite SAMHD1 degradation and high deoxynucleoside triphosphates supply.
Dragin L, Nguyen LA, Lahouassa H, Sourisce A, Kim B, Ramirez BC, Margottin-Goguet F. Retrovirology. 2013 Mar 11;10:30. doi: 10.1186/1742-4690-10-30.
- Identification de HLTF en tant que cible cellulaire dégradée par Vpr du VIH-1 (l’une des premières cibles endogènes de Vpr). HIV-1 Vpr degrades the HLTF DNA translocase in T cells and macrophages.
Lahouassa H, Blondot ML, Chauveau L, Chougui G, Morel M, Leduc M, Guillonneau F, Ramirez BC, Schwartz O, Margottin-Goguet F. Proc Natl Acad Sci U S A. 2016 May 10;113(19):5311-6. doi: 10.1073/pnas.1600485113.
- Découverte d’une nouvelle fonction de Vpx, sa capacité à réactiver les virus latents grâce à la dégradation de HUSH (un rôle inattendu de la protéine virale).
HIV-2/SIV viral protein X counteracts HUSH repressor complex.
Chougui G, Munir-Matloob S, Matkovic R, Martin MM, Morel M, Lahouassa H, Leduc M, Ramirez BC, Etienne L, Margottin-Goguet F. Nat Microbiol. 2018 Aug;3(8):891-897. doi: 10.1038/s41564-018-0179-6.
- Caractérisation de différences entre les mécanismes de dégradation de HUSH et de SAMHD1 par Vpx de HIV-2.
Binding to DCAF1 distinguishes TASOR and SAMHD1 degradation by HIV-2 Vpx.
Martin MM, Matkovic R, Larrous P, Morel M, Lasserre A, Vauthier V, Margottin-Goguet F. PLoS Pathogens 2021 Oct 26;17(10):e1009609. doi: 10.1371/journal.ppat.1009609.
- Caractérisation de différences entre la régulation des facteurs de restriction SAMHD1 et HUSH.
HUSH-mediated HIV silencing is independent of TASOR phosphorylation on threonine 819.
Vauthier V, Lasserre A, Morel M, Versapuech M, Berlioz-Torrent C, Zamborlini A, Margottin-Goguet* F, Matkovic* R. Retrovirology. 2022 Oct 29;19(1):23. doi: 10.1186/s12977-022-00610-7 (* : co-corresponding)
Contact
MARGOTTIN-GOGUET Florence
Team leader, Research director Inserm
Orcid number: 0000-0002-3124-6690
Researcher ID: F-9272-2013
Web of Science: indicate for "author": "Margottin F or Margottin-Goguet F"
Pubmed link : http://www.ncbi.nlm.nih.gov/pubmed?term=Margottin-goguet F[Author] or Margottin F[Author]